An amplicon-sequencing TILLING platform established by ICS-CAAS supporting barley genetic studies
Date:2022-07-12Author:Ping YangSource:
Based on constant efforts over the past five years, the Innovation Team of Wheat and Barley Elite Germplasm Resources Exploration and Innovative Utilization from the Institute of Crop Sciences of the Chinese Academy of Agricultural Sciences (ICS, CAAS) has built up a platform, that combines the chemically-induced genetic resources in conjugation with a high-throughput mutant detection technology, for the functional genomics researches in cultivated barley. The related research have been published in Plant Communications.
The cultivated barley (Hordeum vulgare ssp. vulgare L.) is the fourth most important cereal crop after maize, rice and wheat globally. As a diploid species with a genome smaller than those of other members of the Triticeae tribe, barley has become an attractive model species for the genetic studies in Triticeae crops. The rapid development and significant achievement in barley genomics demand a high-throughput platform to identify the genetically-uniform mutants used for genes functional investigation.
The scientists developed an ethyl methanesulfonate (EMS)-mutagenized population consisting of 8,525 M3 lines in the Chinese barley landrace ‘Hatiexi’ (HTX), which is supplied with a high-quality chromosome-scale assembly of the reference genome. By deployment of multiple state-of-the-art technologies, namely Cel I digestion together with capillary electrophoresis, the whole genome re-sequencing, and sequencing of PCR amplicons, we found that the mutation rate within the population was variable from 1.51 to 4.09 mutations per Mb, depending on the treatment dosage of EMS and the mutation discrimination approaches that applied. In order to establish a highly-efficient and cost-friendly TILLING (Targeting Induced Locus Lesion IN Genomes) platform, a three-dimensional DNA pooling strategy was implemented combining with multiplexed PCR amplicon-sequencing. Mutations were successfully identified from 72 mixed amplicons within a DNA pool containing 64 individual mut
A--nts, and from 56 mixed amplicons within a pool containing 144 individuals. Abundant allelic mutants at the dozens of gene loci, including the barley green revolution contributor gene Brassinosteroid insensitive1 (BRI1) have been revealed. As a proof of concept, the scientists rapidly cloned the causal gene responsible for a chlorotic mutant by applying the MutMap strategy, demonstrating the value of this resource to support forward and reverse genetic studies in barley.
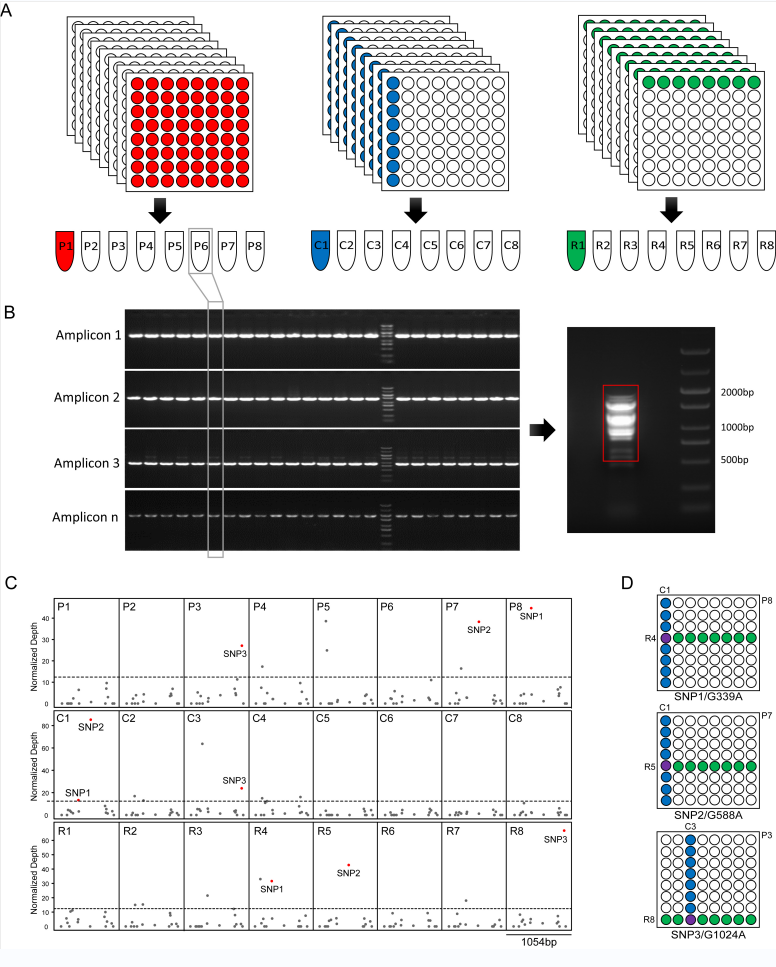
Figure: A TILLING procedure combining the three-dimensional pooling strategy and the amplicon-sequencing based mutants discrimination.
Professor Ping Yang from ICS, CAAS and Dr. Martin Mascher from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK-Gatersleben, Germany) are the corresponding authors. The scientists from Sichuan Agricultural University, and Institute of Botany, Chinese Academy of Sciences, as well as Helmholtz Center Munich/German Research Center for Environmental Health (Munich, Germany), and were involved in collaborations of this work. This work was funded by grants from the National Key Research and Development Program of China (2018YFD1000702/2018YFD1000700) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Reference resource: https://doi.org/10.1016/j.xplc.2022.100317
