RNA Transcript-templated Homology-directed DNA Repair Enables Precise Gene Replacement in Rice
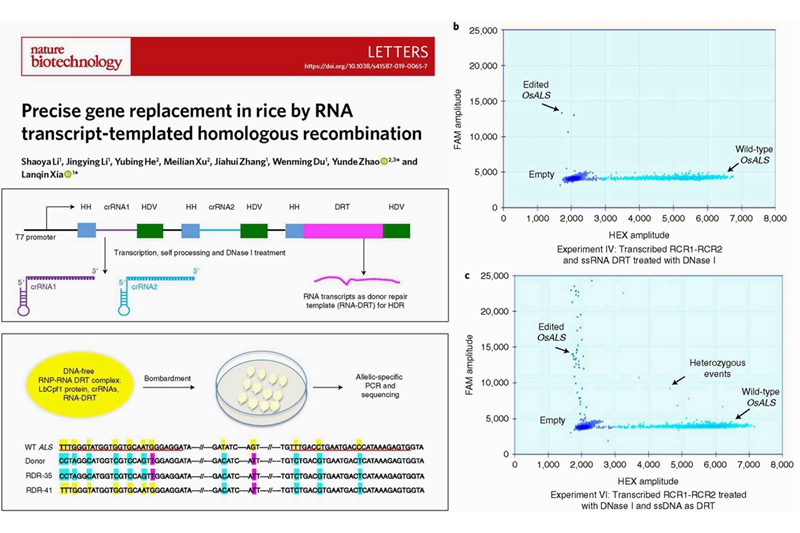
One of the main obstacles to achieve gene replacement in plants is efficient delivery of a donor repair template (DRT) into the nucleus for homology-directed DNA repair (HDR) of double-stranded DNA breaks (DSBs). Production of RNA templates in vivo for transcript-templated HDR (TT-HDR) could overcome this problem, but primary transcripts are often processed and transported to the cytosol, rendering them unavailable for HDR. Recently, by cooperating with University of California, San Diego, the Innovation Team of Crop Transgenesis Technologies and Applications, Institute of Crop Sciences (ICS), Chinese Academy of Agricultural Sciences (CAAS), published a research paper entitled "Precise gene replacement in rice by RNA transcript-templated homologous recombination" in Nature Biotechnology. In this study, the authors showed that coupling Cpf1 (CRISPR from Prevotella and Francisella 1) to an array of CRISPR RNAs (crRNAs) flanked with ribozymes, and a DRT flanked with either ribozymes or crRNA targets, produced primary transcripts that self-process to release the crRNAs and DRT inside the nucleus. Then, they replaced the rice Acetolactate synthase (ALS) gene with a mutated version using a DNA-free ribonucleoprotein (RNP) complex that contains the recombinant Cpf1, crRNAs, and DRT transcripts. They also produced stable lines with two desired mutations in the ALS gene using TT-HDR.
Since the recovery of CRISPR/Cas genome editing technology in 2012, it has been widely used in species such as animals, plants and microorganisms. In general, the double-stranded DNA breaks (DSBs) produce by CRISPR/Cas system are mainly repaired through either non-homologous end joining (NHEJ) or homology-directed DNA repair (HDR). NHEJ is not precise and often used in generating knock-outs and loss-of-function mutants, whereas HDR is precise and can be used for precise gene replacement, knock-in or other modifications. In most species, NHEJ is the predominant pathway, whilst HDR is relatively rare. HDR could be stimulated by the homologous repair templates surrounding DSBs. CRISPR/Cas system and repair templates can be delivered into animal cells more efficiently through electroporation, micro-injection or transfection. In plants, Agrobacterium transformation and particle bombardment are the mainly two methods to deliver CRISPR/Cas system and repair templates into cells. Both particle bombardment with additional free donor fragment and virus-based replicons have been used to increase the availability of DRT, but targeted gene replacement in plants still remains very challenging, which limit the extended application of precision genome editing in improvement of agriculturally important traits.
Unlike DNA repair templates, RNA repair templates can be produced in large amounts by transcription in vivo, which provides more templates for HDR. The authors first delivered ribonucleoprotein (RNP) complexes, which contained recombinant LbCpf1 protein, two crRNAs, and the RNA templates, into rice calli by particle bombardment. Then, they evaluated the feasibility and proved that RNA transcripts could be used as repair template for HDR through allelic-specific PCR and droplet digital PCR (ddPCR). Moreover, by coupling Cpf1 (CRISPR from Prevotella and Francisella 1) to an array of CRISPR RNAs (crRNAs) flanked with ribozymes, and a DRT flanked with either ribozymes or crRNA targets, they produced primary transcripts that self-process to release the crRNAs and DRT inside the nucleus. Finally, by using this tandem single transcript containing both crRNAs and DRT, they successfully generated stable lines with two desired mutations in the ALS gene which confers resistance to ALS-inhibiting herbicides. Furthermore, transgene-free lines with precisely edited ALS loci were recovered following segregation in T1 generation. Collectivelly, they have demonstrated for the first time that RNA transcripts can serve as repair templates for HDR in rice, thereby greatly expanding our ability to improve agriculturally important traits. Their TT-HDR technology makes DNA-free HDR feasible, and provides another boost to the potential commercialization of crops with improved traits using CRISPR-mediated HDR technology.
Shaoya Li, a doctoral student at the Institute of Crop Sciences, CAAS, is the first author of this article. Professor Lanqin Xia from Institute of Crop Sciences and Professor Yunde Zhao from the University of California, San Diego are co-corresponding authors. This work is partly funded by the Ministry of Agriculture and Rural Affairs of China (grant No. 2018ZX0801003B to LX and YH), the Ministry of Science and Technology of China (grant No. 2016YFD0100500 to LX), the Ministry of Agriculture of China (grant No. 2016ZX08010003 to LX), the Central Non-Profit Fundamental Research Funding supported by Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (S2018QY05 to LX)
Link: https://www.nature.com/articles/s41587-019-0065-7
