Creation of Novel Wheat Germplasm of High Resistant Starch Through Genome Editing
Diet-related noninfectious chronic diseases, such as diabetes, coronary heart disease, and certain colon and rectum cancers, are major causes of morbidity and mortality in both developed and developing countries. It is estimated that about 600 million people may suffer from diabetes in the year of 2035. Resistant starch (RS), as a major component of dietary health fiber, refers to any starch or starch products that are not digested and absorbed in the stomach or small intestine, resulting in decreased blood sugar level after human consumption. Cereals high in RS have the potential to lower the risk of those serious noninfectious diseases. A cereal grain with high amylose content (AC) is the source of RS. A positive relationship between RS and AC has been well established. Besides, modified amylopectin (AP) structure with longer amylopectin chain lengths also contributes to RS. Although functional properties of a high RS diet are gaining acceptance as a desirable diet for some consumers, cereal crops that are high in RS are not widely available. Thus, there is an increasing need to develop cereal crops high in RS to meet rapidly growing challenges in nutrition for public health at population level.
Common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is a major staple crop consumed by more than 30% of the world population. It is the main source of cereal-based processed products, such as bread, cookies, pasta and noodles. Wheat consumption is increasing worldwide along with increasing affluence. In general, starch contains two major glucose polymers, amylose and amylopectin, which differ in the degree of polymerization (DP) of glucan chains and in the frequency of branches. In wheat endosperm, starch consists of approximately 70-80% amylopectin and 20-30% amylose. Modification of the starch composition of wheat by increasing its RS content presents an opportunity for a potentially large-scale improvement in public health.
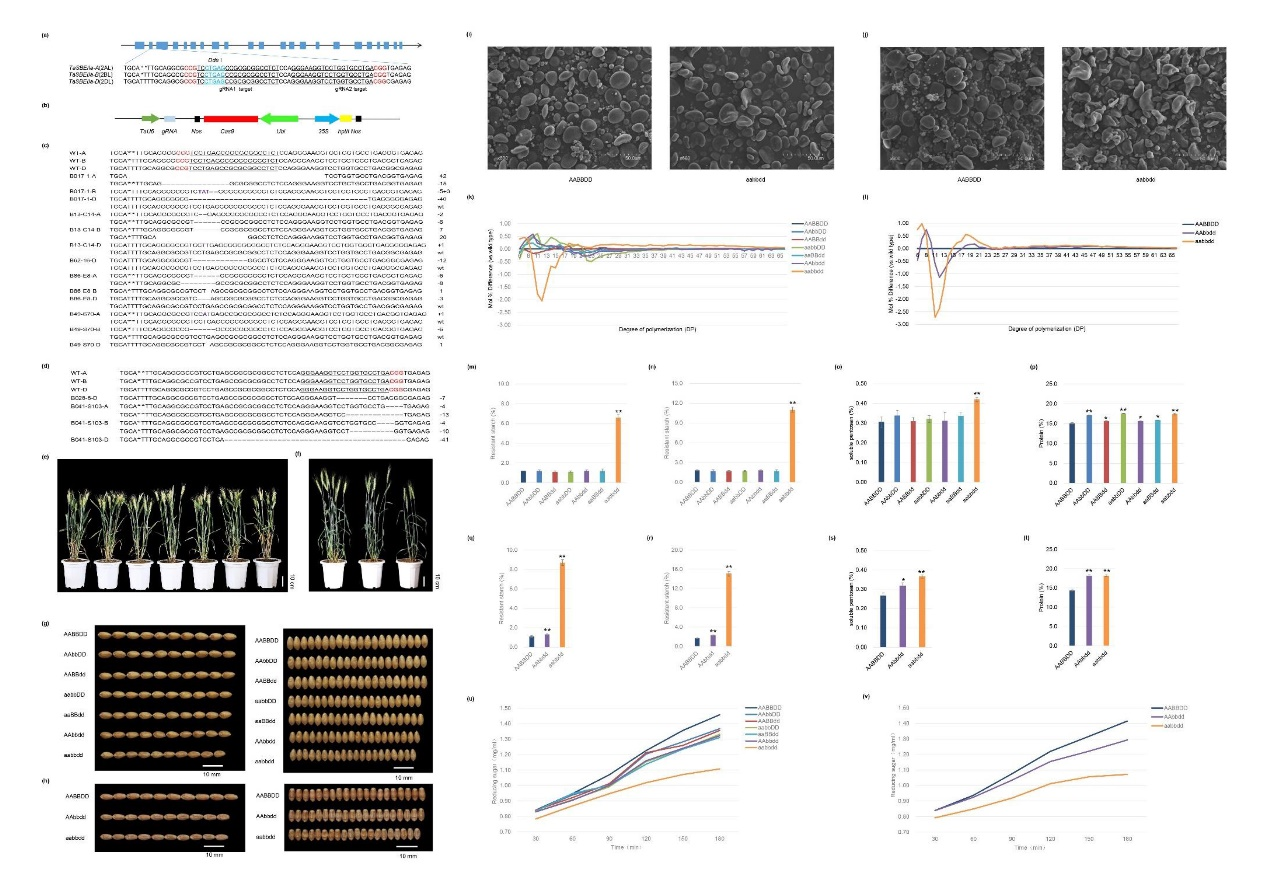
CRISPR/Cas9, as a simple, versatile, robust and cost-effective system for genome manipulation, has recently become the widely used tool for generation of sequence-specific targeted mutagenesis for both functional genomics and crop improvement. The hexaploid nature of common wheat makes generation of targeted mutagenesis in genes, which usually have three homoeologs located in each of the three subgenomes, very challenging. To date, increasing AC and RS contents in wheat through manipulating key genes involved in starch biosynthesis by genome editing has not been documented yet.
Increasing RS in wheat will have a profound impact on global population health. We envisioned that manipulating TaSBEIIa genes through genome editing could be an alternative way to modify the starch composition and structure of wheat to increase its AC and RS contents for human health benefits. In this report, the authors successfully generated a series of transgene-free high amylose wheat plants in both winter wheat cv ZM and spring wheat cv Bobwhite through CRISPR/Cas9-mediated targeted mutagenesis of TaSBEIIa and obtained transgene-free high AC and RS wheat germplasm through genome editing for the first time. They further dissected the roles of three homoeologs of TaSBEIIa in starch composition, amylose content, fine structure of amylopectin, as well as physiochemical and nutritional properties of starch in a sole genetic background. Moreover, they evaluated the effects of different TasbeIIa mutants on end-use qualities including bread- and biscuit-baking qualities. These results provide fundamental information for improving RS content in wheat as well as other cereal crops for global population health benefits.
This work is partly funded by the Ministry of Science and Technology of China (grant no. 2016YFD0100500), the Ministry of Agriculture and Rural Affairs of China (grant no. 2016ZX08010-003), the Central Public-interest Scientific Institution Based Research Fund (Y2020GH04) and National Engineering Laboratory of Crop Molecular Breeding to L.X.
Link: doi.org/10.1111/pbi.13519